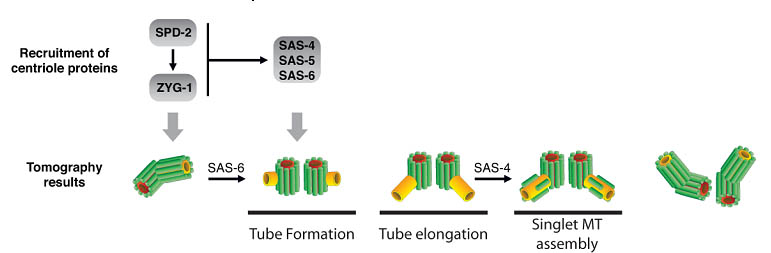
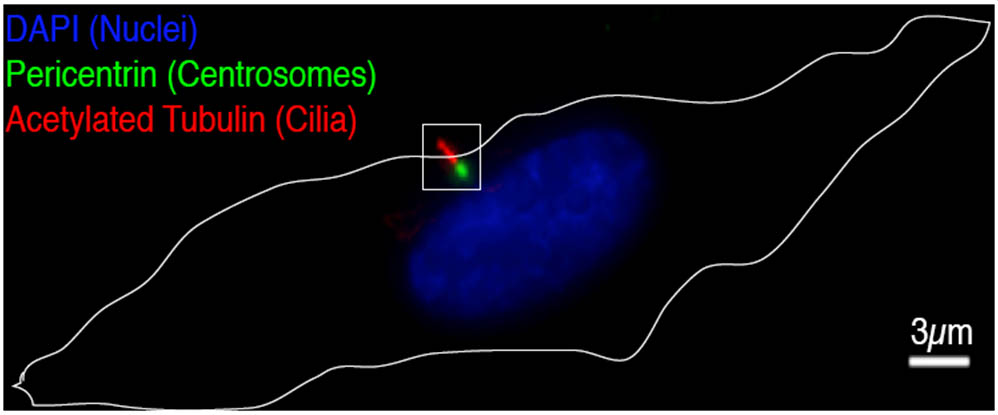
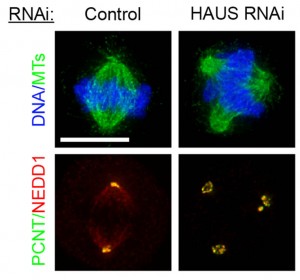
The Pelletier laboratory studies several facets of centrosome biogenesis and function, with a particular emphasis on how their perturbation can lead to devastating developmental diseases and cancer. Centrosomes are composed of a centriole pair surrounded by a proteinaceous matrix composed of over 100 proteins termed pericentriolar material (PCM). By organizing PCM, centrosomes orchestrate several fundamental cellular processes including bipolar mitotic spindle assembly, which plays a determining role in the faithful and accurate segregation of chromosomes to progeny. Centrosome also template the formation of cilia and flagella, key cellular organelles, that play important roles during animal development, cell motility and signaling pathways. Using functional genomic approaches, in combination with cutting-edge microscopy, biochemistry, proteomics and advanced single-cell analytics the overreaching goal of our lab is to identify and study novel proteins required for proper centrosome-cilia biogenesis and function and to investigate their potential roles in human development and disease.
Research in the Pelletier lab revolves around the following axes: